Infer, filter and enhance topological signals in single-cell data using spectral template matching
Goal
Infer, filter and enhance topological signals in single-cell data using spectral template matching
Researchers
- Jonathan Karin
- Yonathan Bornfeld
- Mor Nitzan
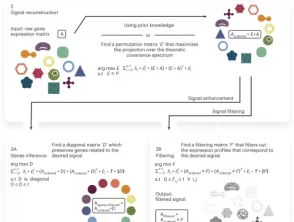
What we did
We apply scPrisma to the analysis of the cell cycle in HeLa cells, circadian rhythm and spatial zonation in liver lobules, diurnal cycle in Chlamydomonas and circadian rhythm in the suprachiasmatic nucleus in the brain. scPrisma can be used to distinguish mixed cellular populations by specific characteristics such as cell type and uncover regulatory networks and cell–cell interactions specific to predefined biological signals, such as the circadian rhythm. We show scPrisma’s flexibility in incorporating prior knowledge, inference of topologically informative genes and generalization to additional diverse templates and systems. scPrisma can be used as a stand-alone workflow for signal analysis and as a prior step for downstream single-cell analysis.
Results
https://www.nature.com/articles/s41587-023-01663-5



